Biological Image Analysis
The goal of this project is identification, spatio-temporal modeling and recognition of dynamical patterns inherent in developmental biology through the use of novel computational tools in image analysis, statistical data aggregation, pattern recognition, machine learning and dynamical modeling. This will lead to the development of new methods for addressing some outstanding challenges in this area, i.e., computing cell lineages, identifying long-term patterns in the tracked output and learning functional models of the dynamics of cell growth and division.
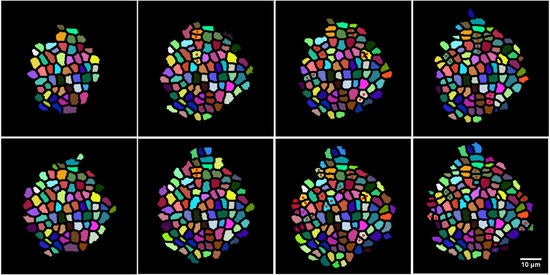
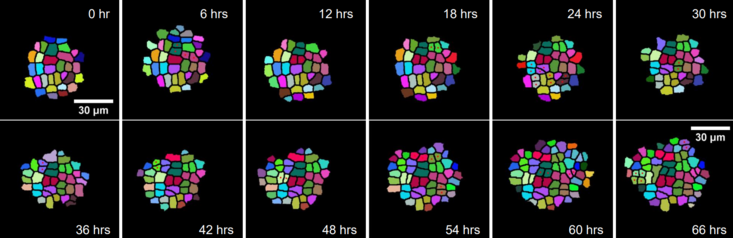
We are working on segmentation, tracking and 3D modeling of cells and tissues in Arabidopsis and Drosophila image stacks. This will lead to the development of new methods for addressing some outstanding challenges in this area, i.e., computing cell lineages, identifying long-term patterns in the tracked output and learning functional models of the dynamics of cell growth and division.
This work has been supported by NSF, and involves collaboration with researchers in Biology and Mathematics.